
Fibroblast growth factor receptor 2 (FGFR2) fusions in intrahepatic cholangiocarcinoma (iCCA)
Genomic alterations in fibroblast growth factor receptors (FGFRs)
- FGFRs are a family of receptor tyrosine kinases.1,2
FGFR signalling pathways play a central role in multiple cellular processes, including cell proliferation, migration and survival1,2
- Alterations in FGFR genes have emerged as tumourigenic drivers in cancers including iCCA, urothelial carcinoma, myeloid/lymphoid neoplasms and other malignancies1,3,4
- FGFR amplifications, mutations and fusions have been observed in all FGFR
subtypes (FGFR1–4).5
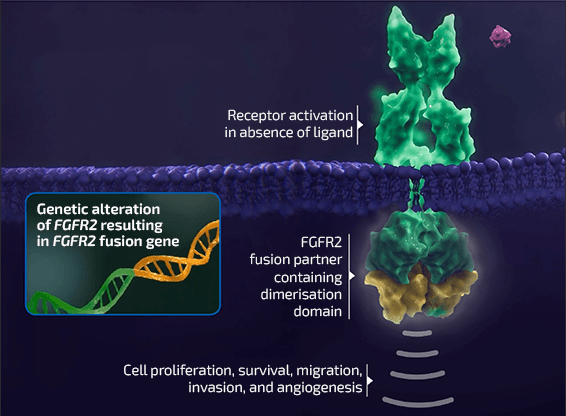
Chromosomal rearrangements involving FGFR2 – resulting in the creation of oncogenic
fusion proteins – have frequently been identified in iCCA6
- Gene fusions are a type of genomic alteration where two independent genes or portions of genes are juxtaposed, resulting in a hybrid gene7,8
- The development of fusion proteins with oncogenic potential can result from gene fusion events involving a range of different partner genes7
FGFR genomic alterations
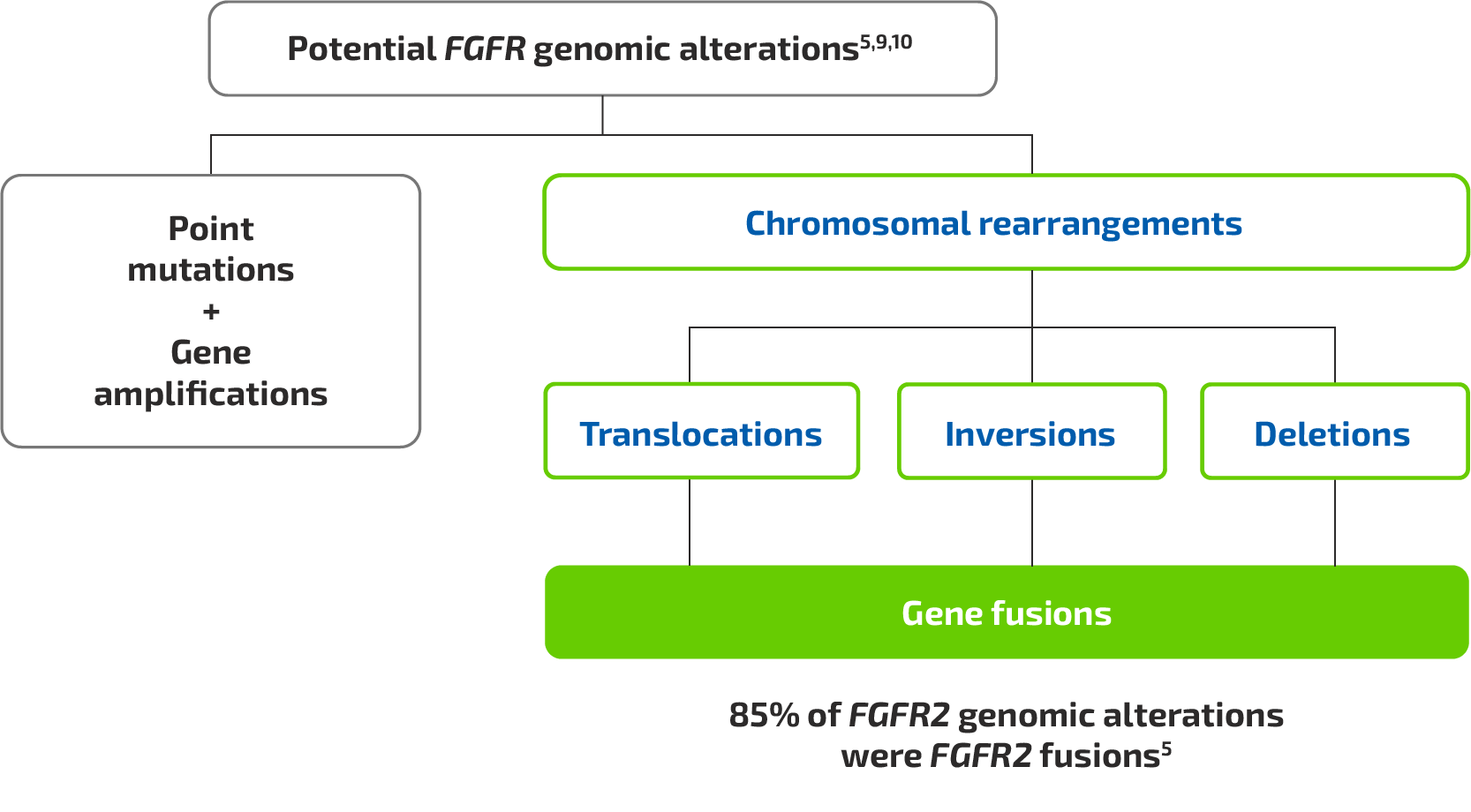
Figure based on Jain A, et al. 2018,5 Lowery MA, et al. 2018,9 and Shibata T, et al. 201810

 Advantages
Advantages
 Challenges
Challenges
